@ratamero YASSSS 🦞 🦀 reminder that the scans are available for all to download on Monash's FigShare instance: https://bridges.monash.edu/articles/dataset/SciPy_2023_Lightning_Talks_Claw/25264588 😃 #SciPy2024
Found not only someone who's also responsible for an OMERO deployment, but who grew up in the tiny English town I lived in for 6 years. #scipy2024 is a small world.
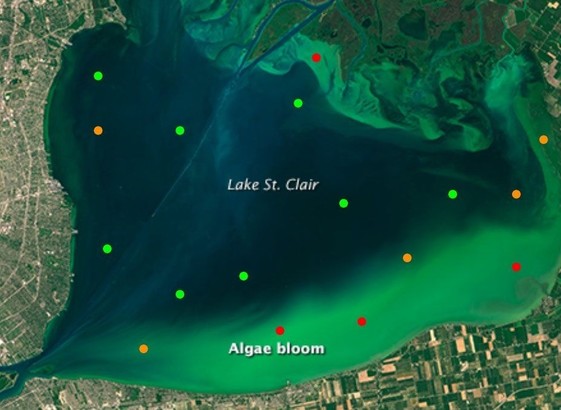
https://cfp.scipy.org/2024/talk/LMF8QH/ If anyone's at #scipy2024 go check out my colleague Emily Dorne's talk today on detecting harmful algal blooms from satellite imagery! Emily took the winning solutions from #drivendata's competition https://www.drivendata.org/competitions/143/tick-tick-bloom , talked to tons of potential users in the natural resource monitoring space, and built a rock solid #python package for anyone to use. An amazing example of how to make impactful #oss
We finally got a @jni shout-out in the lightning talks session (for scanning the claw) #scipy2024
Amazing posters at the poster session at #SciPy2024 🥳
DrivenData data scientist Emily Dorne is speaking today at #scipy2024, discussing cutting-edge developments in Satellite Imagery. Join her at 3pm PST!
Really cool work on solving local problems with satellite imagery and lightweight, effective models.
📢 Anita Sarma, our second keynote of the day, shared her experience about mentoring strategies for an inclusive community, during our diversity luncheon at #SciPy2024 🚀
Thank you for these great insights on mentoring in open source 🫶🏻